
By Aeshah Al-Nagdawi
Highlighting an unprecedented partnership to advance the power of big data in medicine, UC San Francisco has been granted the National Science Foundation’s Convergence Accelerator Award to lead a project in collaboration with data giant Google, Lawrence Livermore National Laboratory, and Institute for Systems Biology. This award testifies to the national importance given to “Harness the Data Revolution” by engaging partnerships between public and private sectors and converging their research efforts.
While a total of twenty-one awards were announced, UCSF is the only one targeting a wide spectrum of biomedicine and health. The award-winning project will be led by Sergio Baranzini, PhD, Professor of Neurology at UCSF, in collaboration with Sharat Israni, PhD, Executive Director and Chief Technology Officer of Bakar Computational Health Sciences Institute.
Called “A Multi-Scale Open Knowledge Network For Precision Medicine,” the project will focus on building a biomedical knowledge engine by synthesizing data from a large number of databases. As an open source, its roll-out will give physicians unprecedented patient care insights and researchers the tools for deeper biomedical inquiries and new drug discovery.
This is the engine at the core of our efforts. This is how we will revolutionize health, research, and clinical care.
Keith Yamamoto, PhD
Vice Chancellor of Science Policy and Strategy and Director of UCSF Precision Medicine
“Drug discovery is where genotypic and phenotypic work converge,” said Israni, who leads the Information Commons and Knowledge Network projects at Bakar Computational Health Sciences Institute. “A wealth of insights are waiting to be discovered.” He cites the example of antipsychotics, which arose from antihistamines. Yet antipsychotics may be acting via many more targets than antihistamines do. Expanding a shared knowledge network opens up the possibility to find these new targets.
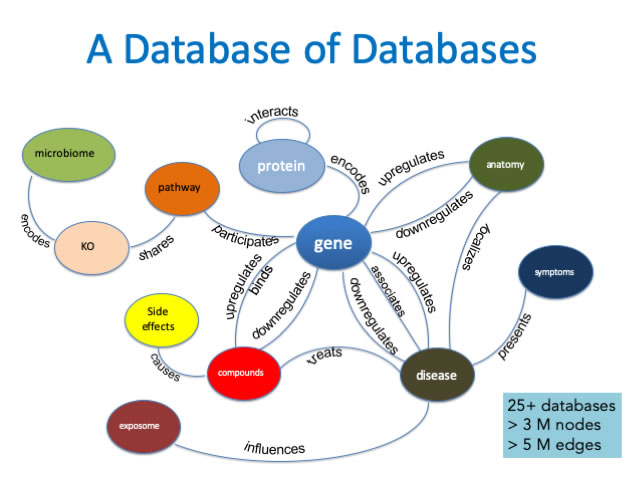
The platform for the new project is UCSF’s Scalable Precision Medicine Knowledge Engine (SPOKE), a network which combines a growing list of databases (25+), linking millions of compounds and tens of thousands of genes, proteins, diseases, and other data. SPOKE allows researchers to explore biomedical structures and their interconnected pathways. Drug discovery and repurposing, and clinical trial cohort characterization are examples of its potential. “SPOKE could hold the key to understanding pathophysiology to the degree that we may some day invent personalized drugs,” Israni said.
The proposed expansion of SPOKE will leverage UCSF’s vast, de-identified clinical data, along with continuous acquisition of new information from lab experiments, clinical trials, and electronic health records from other centers to frame testable hypotheses and create individualized, high-dimensional profiles at unparalleled scope and depth. "Cross-referencing information has always been regarded as a powerful strategy for discovery of new knowledge. SPOKE embodies that concept in biomedicine," said Baranzini.
Unprecedented Partnership
The new partnership among teams of researchers from academia, government, non-profit, and commercial sectors will converge a series of distinct but related projects to create a powerful, open knowledge network, holding promise for a breakthrough impact in the biomedical world. Lawrence Livermore National Labs will undergird the work with their enormous computing capability and innovative methods to explore extremely large networks. ISB will leverage their systems-level analyses of biological processes. Google will oversee the build of a search engine to access UCSF’s dynamic, ever-growing data set from SPOKE.
The outcome of this first phase of the project will be a dramatic expansion of the SPOKE meta-database, available via Google’s datacommons.org, with APIs that enable integration of individual datasets into the network by any contributing party. In a second phase, the team plans to implement the first APIs to make use of the power of SPOKE. One such application will embed deidentified patient data (from UCSF information commons) onto SPOKE to create unique profiles that will characterize patients on a multidimensional platform, allowing for cohort creation and outcome prediction, among other features.
The ultimate goal is to create a single resource for existing and future biomedical knowledge that could be freely used by physicians in the presence of patients and by researchers for a fresh round of discoveries.
“Imagine combining – on a single computational platform – all findings about normal and aberrant biological processes, from human populations, to individuals, to experimental organisms, to cells and molecules,” said Keith Yamamoto, PhD, Vice Chancellor of Science Policy and Strategy and Director of UCSF Precision Medicine. “This is the engine at the core of our efforts. This is how we will revolutionize health, research, and clinical care.”
